Organizing neuroimaging data
We organize our neuroimaging data in BIDS (Brain Imaging data structure) format for better accessibility and sharing across different projects. BIDS was heavily inspired by the format used internally by the OpenfMRI repository that is now known as OpenNeuro. The official website explains the need of BIDS format as follows:
Neuroimaging experiments result in complicated data that can be arranged in many different ways. So far there is no consensus how to organize and share data obtained in neuroimaging experiments. Even two researchers working in the same lab can opt to arrange their data in a different way. Lack of consensus (or a standard) leads to misunderstandings and time wasted on rearranging data or rewriting scripts expecting certain structure. With the Brain Imaging Data Structure (BIDS), we describe a simple and easy to adopt way of organizing neuroimaging and behavioral data.
Our scripts can be found at our Github link: https://github.com/gvbhalerao591
TDCS simulation procedures using MATLAB scripts
We have developed our in-house scripts to easily implement various steps such as segmentation, segmentation post-processing, electrode placement, finite element model generation and solving using MATLAB, ABAQUS/CAE and ScanIP. We have also written custom MATLAB scripts to post-process the simulation outputs in standard brain template which can later be used to calculate the electric field values for different regions of interest. These scripts will cover end-to-end workflow for simulation of TDCS using structural brain MRI. Our scripts can be found at our GitHub link: https://github.com/gvbhalerao591
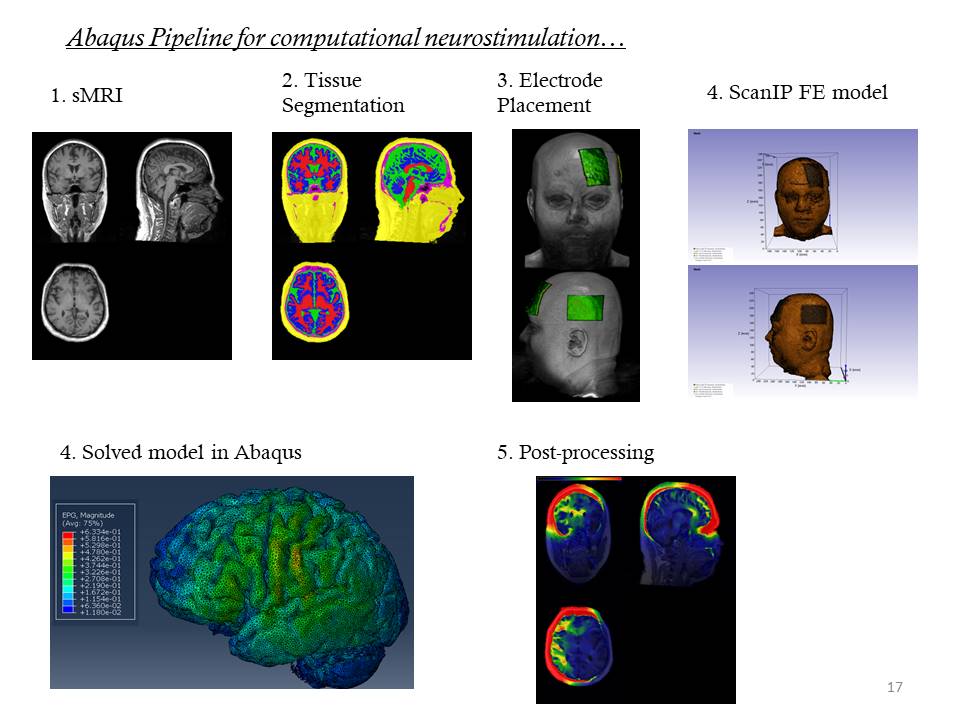
SOPs to implement TDCS modeling pipelines
Since various TDCS modeling tools are available in research community, it is interesting to see how the electric field outputs produced by these pipelines differ. We have used five different TDCS simulation pipelines (ABAQUS, ROAST, SimNIBS). The steps of procedure to implement all these pipelines are provided in a comprehensive document below.
Utilizing multiple (open source & commercial) softwares
We use multiple tools and softwares to implement our pipelines and perform statistical analyses. Our work constantly involves keeping up with the advancements of the softwares in the research community. We have created and generated our standard steps of procedures for end-to-end implementation of structural, resting state and diffusion MRI analyses. To access these details, please email to gvbhalerao591@gmail.com

